Aplacophoran mollusc biodiversity and systematics
|
Aplacophora (Figure 1) is a diverse clade of shell-less, worm-shaped marine molluscs (reviewed by Todt et al. 2008, Todt 2013). Several species are ecologically important (Scheltema 1997) and Aplacophora plus Polyplacophora (chitons) make up the sister taxon of all other molluscs (reviewed by Kocot 2013, Giribet 2014). The number of scientists studying aplacophorans has declined in recent years, leaving just a handful of expert researchers worldwide. Meanwhile, almost every time an expert samples a new place, new species are discovered (e.g., Scheltema 1999, Kocot and Todt 2014, Passos et al. 2016) and both known but undescribed species and unidentified specimens collected in environmental surveys (e.g., Schiaparelli et al. 2014) continue to grow in number. Identifying most aplacophorans has traditionally required the destructive process of histology. However, use of micro-computed tomography (micro-CT) scanning, which enables non-destructive visualization of internal anatomy from several specimens simultaneously, would significantly accelerate the pace of specimen identification. Identification of specimens using molecular approaches would also reduce challenges to studying aplacophoran biodiversity and make the group more accessible to non-experts, but this is hindered by a paucity of available DNA barcodes. Because the group holds an important place in molluscan phylogeny and is morphologically variable in terms of many key molluscan characters (reviewed by García-Álvarez and Salvini-Plawen 2007), understanding the evolutionary polarity of these characters in Aplacophora could also shed light on the early evolution of Aculifera and Mollusca as a whole. Unfortunately, phylogenetic analyses to date have never had representative taxon sampling for Aplacophora (reviewed by Todt 2013).
|
This research is funded by an NSF CAREER grant. The project aims to address fundamental questions about aplacophoran biodiversity and systematics such as: How many species of aplacophorans are there? How widely are species distributed? How rare or common are most species? Does the current taxonomy of the group reflect its evolutionary history? What can improved understanding of the evolutionary polarity of aplacophoran character states tell us about the last common ancestors of Aplacophora, Aculifera, and Mollusca as a whole? To this end, the PI and his team will employ a novel workflow that collects data using light microscopy, micro-CT, scanning electron microscopy (SEM), and DNA barcoding – all from the same specimen. Further, genome and transcriptome sequencing, target-capture phylogenomics, ancestral character state reconstruction, and molecular clock analyses will be performed to provide a robust and broadly sampled phylogenetic framework, a revised classification system for Aplacophora, and new insight into the early evolution of Mollusca in light of recent phylogenomic work (Figure 2). The specific objectives of this proposal are to:
- Address the erosion of taxonomic expertise for many small benthic marine invertebrate taxa by hosting workshops to integrate training of the next generation of invertebrate systematists with essential specimen collection.
- Use an integrative taxonomic approach to identify and describe species, prepare monographs on groups that are particularly understudied or in need of revision, characterize the faunas of select understudied regions, and aid future researchers in identifying specimens by producing a library of DNA barcodes from expert-identified specimens.
- Reconstruct aplacophoran phylogeny using a phylogenomic approach with broad taxon sampling and revise aplacophoran taxonomy to reflect its phylogeny.
- Conduct ancestral character state reconstruction and molecular clock analyses to infer the plesiomorphic states of key morphological characters and timing of major events in molluscan evolution, thus shedding light on the early evolution of the most diverse animal phylum in the sea.
|
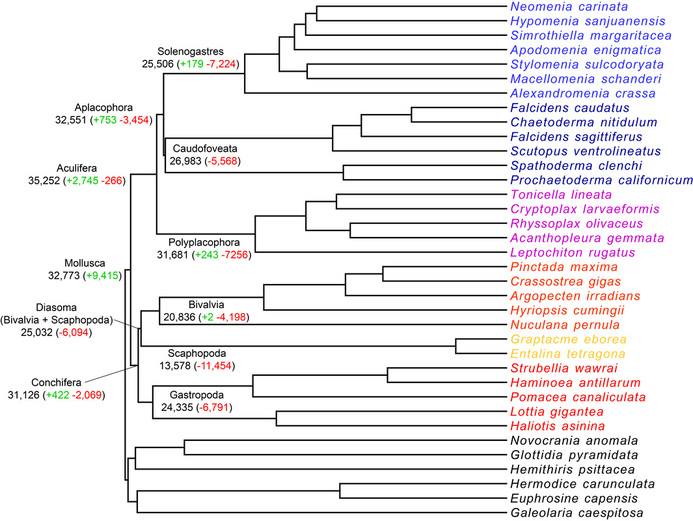
Figure 2. Higher-level molluscan phylogeny based on recent a phylogenomic study (Kocot et al. 2019). |
Scaphopod genomics and phylogenetics
|
Scaphopoda (tusk shells) is a relatively small class of Mollusca with an incompletely understood evolutionary history. Relationships within the group are poorly known. Moreover, the placement of Scaphopoda with respect to Gastropoda and Bivalvia are also uncertain. Traditional morphology-based views placed Scaphopoda + Bivalvia in a clade called Diasoma (Runnegar and Pojeta 1974). However, recent molecular studies have recovered Scaphopoda as sister to Gastropoda (Smith et al. 2011) or sister to Gastropoda + Bivalvia (Kocot et al. 2011, Vinther et al. 2012). In a fashion similar to what is described for the aplacophorans above, we are planning on employing a target capture approach to take advantage of collections from Iceland and Antarctica (Figure 3) as well as museum specimens to build a large-scale, robustly resolved phylogeny for Scaphopoda.
|
Figure 3. Siphonodentalium dalli female (top) and male (botom) collected from Antarctica during the IcyInverts 2020 cruise (NBP 20-10). Photo by Nick Roberts.
|
Evolutionary genomics of biomineralization in molluscs
Biomineralization is the process by which living organisms convert ions in solution into solid minerals (Simkiss and Wilbur 1989). The great success of molluscs can be attributed in part to their ability to secrete calcareous skeletal structures with fossil evidence for biomineralization in molluscs extending back to the late Precambrian (Runnegar 1996). This remarkable capacity has evolved to produce a phenomenal range of skeletal structures with a diversity of shapes and properties. The ability of molluscs to fabricate these intricate and robust structures from sea water, which is encoded in their genomes, is well beyond human engineering capabilities. While there has been progress in identifying and characterizing genes involved in biomineralization (Jackson et al. 2006, 2010, Marie et al. 2010, Zhang et al. 2012), there remain fundamental gaps in our understanding of this process. For instance, it is known that dramatically different repertoires of proteins pattern mineralized structures in different molluscan taxa (Jackson et al. 2010), but the biomineralization toolkits present in most molluscan lineages are yet to be determined. Likewise, little is known about the evolutionary history and origins of biomineralization genes but it appears that both evolutionarily conserved genes (Le Roy et al. 2014) as well as rapidly evolving genes (McDougall et al. 2013) are involved.
The objectives of this work are to employ new genomic resources and a well-resolved phylogeny for Mollusca to identify biomineralization genes in important but understudied lineages and infer the evolutionary history of these genes using an evolutionary genomic approach. Three aims are proposed:
Researchers in the fields of materials science, biomimicry, and synthetic biology are interested in reproducing biomineralization in vitro because of valuable biomedical (among other) applications (Luz et al. 2009, Meyers et al. 2013). However, the genetic toolkit responsible for this process and how it evolved must first be understood. Studies of molluscan biomineralization are also important to climate change research as shells of certain ecologically important snails are already being dissolved by ocean acidification (Hoffmann et al. 2010). Further, understanding of the genetics and evolutionary history of biomineralization is important to the fields of invertebrate biology, evolutionary developmental biology and paleontology (e.g., Aguilera et al. 2014, Vinther 2015) as this would help answer important questions about the evolution of animal body plans and the genetic toolkits that underlie them (Figure 4).
Although there is great interest in understanding evolution of molluscan biomineralization, the vast majority of studies to date have been conducted on gastropods and bivalves (the only molluscs with published genomes) whereas biomineralization in other lineages has received virtually no attention. Understanding of molluscan biomineralization is also hindered by lack of a resolved phylogenetic framework for the group. Despite recent efforts (e.g., Kocot et al. 2011, Smith et al. 2011), two important aspects of the mollusc tree of life remain unresolved, hindering evolutionary studies on the group in general.
The objectives of this work are to employ new genomic resources and a well-resolved phylogeny for Mollusca to identify biomineralization genes in important but understudied lineages and infer the evolutionary history of these genes using an evolutionary genomic approach. Three aims are proposed:
- Resolve evolutionary relationships among the conchiferan molluscs, producing a robust phylogenetic framework onto which the evolution of biomineralization genes will be traced.
- Identify the biomineralization gene repertoires in representatives of key lineages of Mollusca using whole genome sequencing and infer their evolutionary history using an evolutionary genomics approach.
- Use single-cell transcriptomics and proteomics to examine gene expression and protein localization to gain insight into the function of biomineralization genes.
Researchers in the fields of materials science, biomimicry, and synthetic biology are interested in reproducing biomineralization in vitro because of valuable biomedical (among other) applications (Luz et al. 2009, Meyers et al. 2013). However, the genetic toolkit responsible for this process and how it evolved must first be understood. Studies of molluscan biomineralization are also important to climate change research as shells of certain ecologically important snails are already being dissolved by ocean acidification (Hoffmann et al. 2010). Further, understanding of the genetics and evolutionary history of biomineralization is important to the fields of invertebrate biology, evolutionary developmental biology and paleontology (e.g., Aguilera et al. 2014, Vinther 2015) as this would help answer important questions about the evolution of animal body plans and the genetic toolkits that underlie them (Figure 4).
Although there is great interest in understanding evolution of molluscan biomineralization, the vast majority of studies to date have been conducted on gastropods and bivalves (the only molluscs with published genomes) whereas biomineralization in other lineages has received virtually no attention. Understanding of molluscan biomineralization is also hindered by lack of a resolved phylogenetic framework for the group. Despite recent efforts (e.g., Kocot et al. 2011, Smith et al. 2011), two important aspects of the mollusc tree of life remain unresolved, hindering evolutionary studies on the group in general.
|
Figure 4. Preliminary gene family evolution analysis showing gain and loss of secreted protein gene families in various clades of molluscs. Ancestrally, Mollusca appears to have had 32,773 genes encoding proteins with a signal peptide including 9,415 unique to molluscs (with respect to sampled outgroups). However, because these taxa are mostly represented by transcriptomes and not genomes, this preliminary analysis likely overestimates losses and underestimates gains. |
Lophotrochozoan phylogeny
The evolutionary relationships among Lophotrochozoa, the clade that includes animals such as molluscs, annelids, flatworms, bryozoans, and others, have long been an enigma. See Kevin's review article on the topic. Ongoing work in the lab seeks to resolve higher-level lophotrochozoan phylogeny using a phylogenomic approach as well as non-traditional sources of data (e.g., rare genomic changes). Ph.D. student Nick Roberts is leading work that will greatly expand the taxon sampling of Lophotrochozoa relative to previous studies and screen these and publicly available data for contamination, which may have introduced misleading signal in previous studies.
Dragonfly conservation genomics and eDNA
In collaboration with Dr. John Abbott and Kendra Abott, we are investigating the distribution, population genetics, and ancestral population sizes of imperiled odonates from the Southeastern United States. This work will employ use of environmental DNA (eDNA) to detect juvenile dragonflies, which can be very cryptic, in their natural habitat in a non-destructive way. Whole genome sequencing of select species will be used to infer ancestral population size using pairwise Markov sequential coalescent approaches. This work is funded by the Texas Parks and Wildlife Division and Louisiana Wildlife and Fisheries.
Phylogeography of deep-sea Icelandic invertebrates
In 2011, Kevin participated in the IceAGE Proect. IceAGE aims to combine classical taxonomic methods with modern aspects of biodiversity research, in particular phylogeography (population genetics and DNA barcoding) and ecological modelling in the sensitive region around Iceland. The sampling area is characterized by several local peculiarities like submarine ridges (geographical barriers) and influence of different water masses of different origin. This allows the analysis of factors influencing the distribution and migration of species as well as investigation of the background of biogeographic zonation.
Other interests
In no particular order, here's some other topics/taxa we are interested in:
- Meiofauna
- Entoprocts
- Hemichordates
- Kinorhynchs
- Peracarid crustacean phylogeny
- Bopyrid isopods
- 'Mesozoans'
- Methods in orthology inference